___
Grape R2R3-MYB Family
-We have revisited the grape R2R3 MYB gene family (Wong et al., 2016) and adopted their names according to the Super Nomenclature Committee.
–Phylogenetic Trees
The following trees show very similar topologies and clustering of functional subgroups described by Kelemen et al., 2015.
1. Phylogenetic relationships between Vitis and Arabidopsis R2R3-MYB families. (constructed by JTM in Wong et al., 2016).
Grapevine accessions shown with asterisks correspond to RNA-Seq-confirmed or improved sequences. Proteins isolated from other cultivars are shown with letters (CS: Cabernet Sauvignon, SH: Syr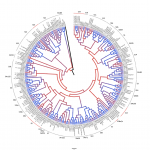 ah). Annotations without RNA-Seq confirmation were taken from the Pinot Noir reference genome (PN). AtmaMYB was used as outgroup (AT5G45420). Red lines represent subgroups that were further defined based on the presence of C-terminal motifs identified in MEME and/or described by Stracke et al. (2001). Bootstrap values (>75) are shown on a coloured scale ranging from purple to blue. Click on thumbnail to access the high-resolution image. You can also access the PDF version (no subgroup numbering) for manually inspecting IDs, click here.
ah). Annotations without RNA-Seq confirmation were taken from the Pinot Noir reference genome (PN). AtmaMYB was used as outgroup (AT5G45420). Red lines represent subgroups that were further defined based on the presence of C-terminal motifs identified in MEME and/or described by Stracke et al. (2001). Bootstrap values (>75) are shown on a coloured scale ranging from purple to blue. Click on thumbnail to access the high-resolution image. You can also access the PDF version (no subgroup numbering) for manually inspecting IDs, click here.
2. Bayesian Tree for Maximum Likelihood Vitis-Arabidopsis using AQUILO (UniProtKB – D7T987) as outgroup (constructed by Alberto Perez-Tejeda, Master’s degree student in Bioinformatics).
Grapevine accessions show the cultivars in which these sequences were taken (PN: Pinot Noir, CV: Corvina, ML: Merlot) and the A. thaliana were taken from The Arabidopsis Informati
___
Cannabis R2R3-MYB Family
–Phylogenetic Trees
Constructed by Alberto Perez-Tejeda, Master’s degree student in Bioinformatics.
1. At+Cs Tree.
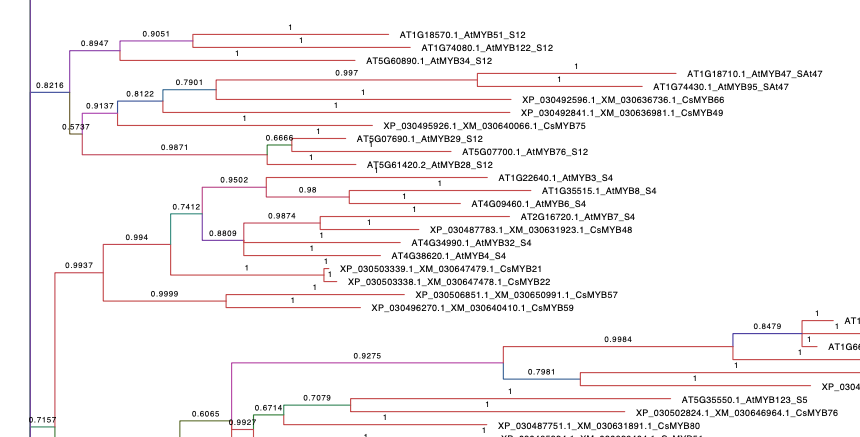 Refers to the phylogenetic tree containing A. thaliana and C. sativa R2R3 MYB family. A. thaliana sequences were taken from The Arabidopsis Information Resource (TAIR) database and C. sativa MYBs present the same name as the published by (Bassolino et al., 2020). The alignment of the sequences was performed with MUSCLE (parameters by default). By using MEGA X, the best protein model (ML), searched based on the lowest Bayesian information criterion (BIC) score, corresponded to the Jones-Taylor-Thornton (JTT) substitution model and rates among sites following a gamma distribution (G). This analysis was carried on MrBayes software and 12.5 millions generations were performed with a final standard deviation(sd)=0.098034. Numbers in branches display the probability of a correct tree according to our data.Branches are coloured according to their probability.
Refers to the phylogenetic tree containing A. thaliana and C. sativa R2R3 MYB family. A. thaliana sequences were taken from The Arabidopsis Information Resource (TAIR) database and C. sativa MYBs present the same name as the published by (Bassolino et al., 2020). The alignment of the sequences was performed with MUSCLE (parameters by default). By using MEGA X, the best protein model (ML), searched based on the lowest Bayesian information criterion (BIC) score, corresponded to the Jones-Taylor-Thornton (JTT) substitution model and rates among sites following a gamma distribution (G). This analysis was carried on MrBayes software and 12.5 millions generations were performed with a final standard deviation(sd)=0.098034. Numbers in branches display the probability of a correct tree according to our data.Branches are coloured according to their probability.
2. Cs+Vvi Tree.
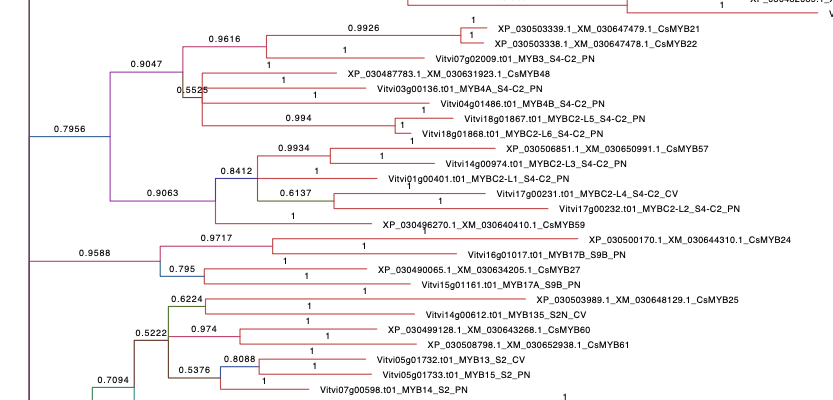 Refers to the phylogenetic tree containing C. sativa and V. vinifera R2R3 MYB family. Grapevine accessions show the cultivars of origen (PN: Pinot Noir, CV: Corvina, ML: Merlot) and C. sativa MYBs present the same name as the published by (Bassolino et al., 2020). The alignment of the sequences was performed with MUSCLE (parameters by default). By using MEGA X, the best protein model (ML), searched based on the lowest Bayesian information criterion (BIC) score, corresponded to the Jones-Taylor-Thornton (JTT) substitution model and rates among sites following a gamma distribution (G). This analysis was carried on MrBayes software and 14 millions generations were performed with a final standard deviation(sd)=0.097856. Numbers in branches display the probability of a correct tree according to our data.Branches are coloured according to their probability.
Refers to the phylogenetic tree containing C. sativa and V. vinifera R2R3 MYB family. Grapevine accessions show the cultivars of origen (PN: Pinot Noir, CV: Corvina, ML: Merlot) and C. sativa MYBs present the same name as the published by (Bassolino et al., 2020). The alignment of the sequences was performed with MUSCLE (parameters by default). By using MEGA X, the best protein model (ML), searched based on the lowest Bayesian information criterion (BIC) score, corresponded to the Jones-Taylor-Thornton (JTT) substitution model and rates among sites following a gamma distribution (G). This analysis was carried on MrBayes software and 14 millions generations were performed with a final standard deviation(sd)=0.097856. Numbers in branches display the probability of a correct tree according to our data.Branches are coloured according to their probability.
