Tomato (Solanum lycopersicum cv. ‘Heinz’)
Silkworm Mulberry (Morus alba)
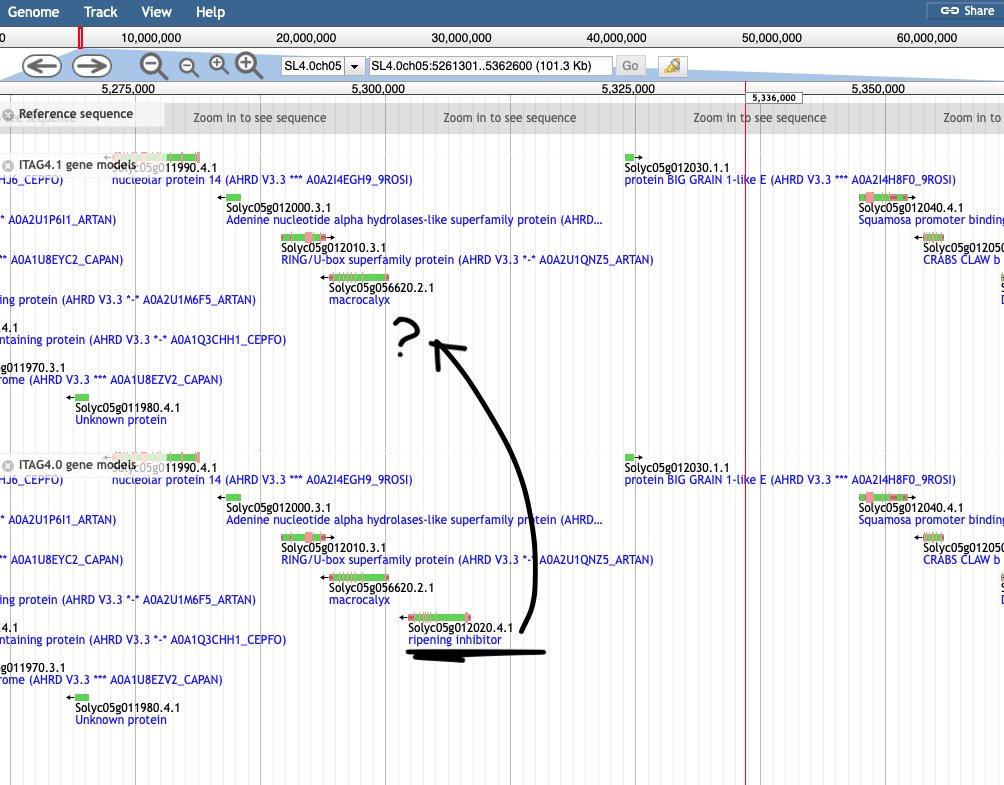
We have been updating the Morus alba genome annotation gff with our genes of interest. The original article is from Jiao et al. (2020)
Ut convallis, magna sed dapibus tincidunt, nulla lacus sollicitudin nisi, id commodo urna urna in elit. Nunc vulputate tincidunt risus non.


